Infections from antibiotic resistant bacteria can be treated with small proteins found in the microbiome according to a new study.
By Swetha Kasetty Ramakrishnan
“Superbugs” are bacteria that are getting harder to kill because they resist multiple currently used antibiotics. Imagine if proteins found in our bodies could be the key to beating these superbugs. Scientists are doing just that by studying the human microbiome—a collection of microbes that live in and on us.
In recent years, many common infections have become tougher to treat due to antibiotic resistant bacteria. This means that some bacteria don’t respond to the usual medicines anymore, making it hard for doctors to treat infections. Superbugs are resistant to numerous antibiotics; this can cause major health issues. Scientists are on a quest to find new solutions. One promising area of research involves proteins called antimicrobial peptides (AMPs).
RELATED: Searching for Alternatives to Antibiotics
Antimicrobial peptides (AMPs)
AMPs are natural defenders against bacteria. They are found in many living things, from single-celled organisms to humans. AMPs have the advantage of being small and having the ability to attack bacteria in various ways. Our microbiome already has AMPs that help protect us from harmful bacteria. Scientists in a recent study from the labs of the University of Pennsylvania School of Engineering and Applied Science and Stanford University took the large database of small proteins from our microbiome to find those with the potential to fight infections from antibiotic resistant bacteria.
RELATED: A Cancer Treatment Using Patient’s Own Immune System
Finding AMPs
The researchers started with a large dataset of predicted AMPs taken from samples of the human microbiome. However, they chose to look for smORF-encoded peptides (SEPs). SEPs are different compared to previously identified AMPs. SEPs kill bacteria by attacking their membrane, making holes. This leads to cell death. They can work together in a process called synergism, where the combined effect of two or more small proteins is greater than the sum of their individual effects. Additionally, SEPs can influence gut commensals—beneficial bacteria living in the gut—by helping maintain a balanced microbial community. This modulation can ensure that harmful bacteria are kept in check while supporting the growth of beneficial ones, contributing to overall gut health.
The scientists wrote computer programs to predict which of the proteins might be used as antibacterials to treat antibiotic resistant bacteria. After narrowing down the list, they synthesized, or made, 78 of these peptides in the lab. Then, they tested them against different harmful bacteria. The most promising AMPs were found in different areas of the body, like the mouth, skin, and gut. One standout, prevotellin-2, was a particularly effective treatment for mice infected with harmful bacteria while being safe for the mice.
What does this mean for antibiotic resistant bacteria?
If the solution to treat superbugs was always within us, why has it taken so long for scientists to look for them? First, we have only recently generated large microbiome databases. Second, their small size makes finding them challenging. Third, complicated computer programs are required to look through the data and identify the proteins. Lastly, identifying the proteins doesn’t guarantee their effectiveness. The proteins must be artificially made and tested.
In summary, the findings highlight the potential of AMPs fighting bacterial infections. While the study has some limitations, like not identifying all small proteins, it lays a strong foundation for future studies. By continuing to explore AMPs, scientists may discover new ways to develop safe and effective treatments against infections from antibiotic resistant bacteria.
This study was published in the peer-reviewed journal Cell.
Reference
Torres, M. D. T., Brooks, E. F., Cesaro, A., Sberro, H., Gill, M. O., Nicolaou, C., Bhatt, A. S., & de la Fuente-Nunez, C. (2024). Mining human microbiomes reveals an untapped source of peptide antibiotics. Cell, 187(19), P5453-5467.E15. https://doi.org/10.1016/j.cell.2024.07.027
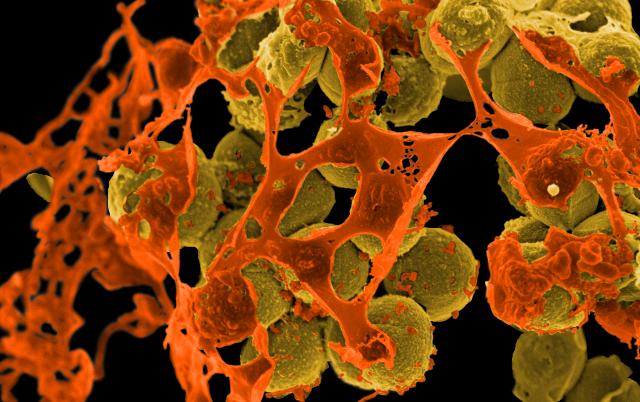
Featured image: Scanning electron micrograph of methicillin-resistant Staphylococcus aureus (MRSA, yellowish-brown) surrounded by cellular debris. MRSA resists treatment with many antibiotics. Credit: Methicillin-Resistant Staphylococcus aureus (MRSA) Bacteria by NIAID is licensed under CC BY 2.0.
The information contained in this article is for informational purposes only and is not intended as health or medical advice. Always consult a physician or other qualified health provider regarding any questions you may have about a medical condition or health objectives.
About the Author
Swetha Kasetty Ramakrishnan has a PhD in microbiology from Dartmouth College. She is currently a budding science communicator and enjoys sharing a good science story with anyone willing to listen. She hopes to make science more accessible and approachable to everyone.